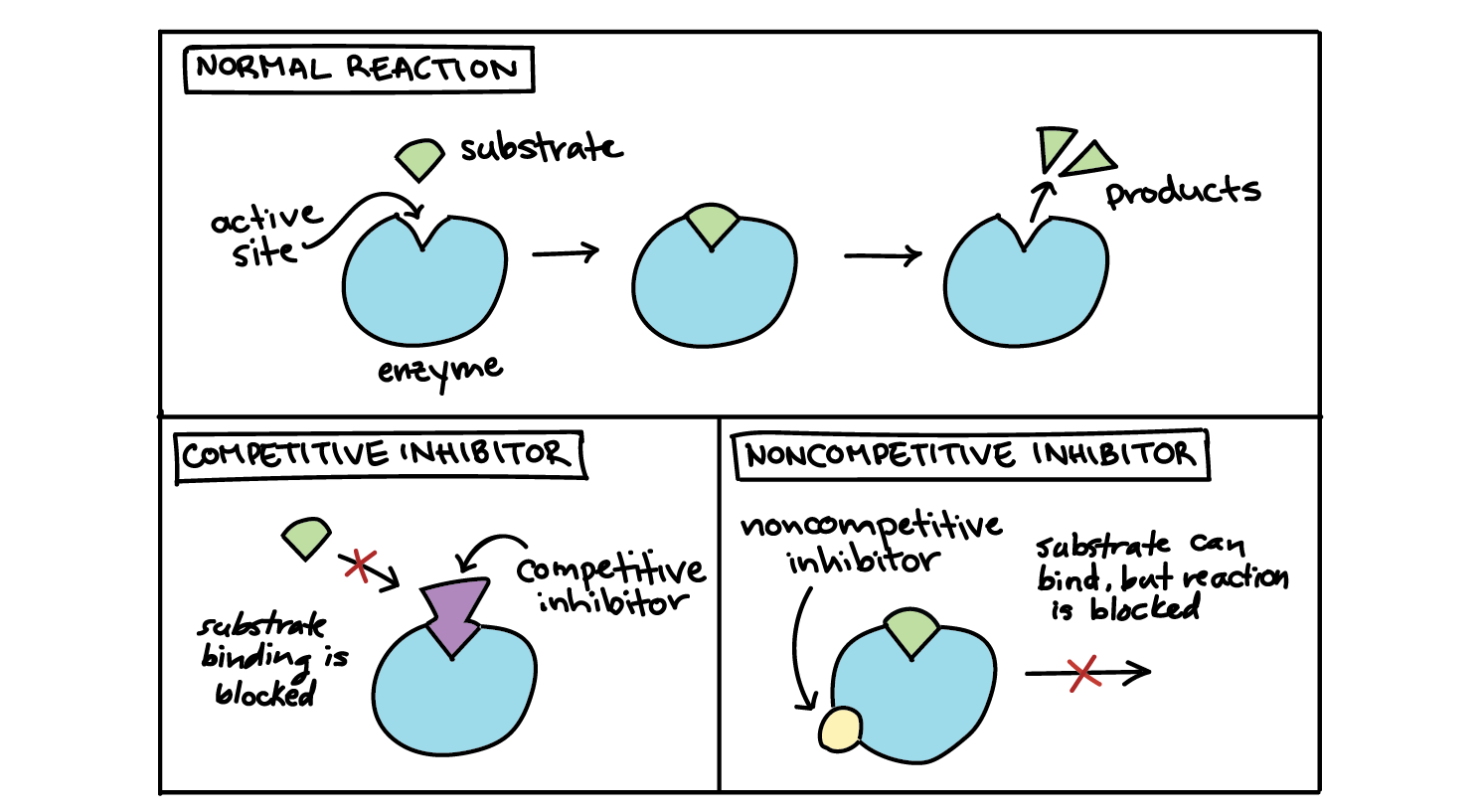
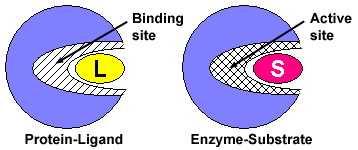
Representation of substrate binding to the active site of an enzyme... | Download Scientific Diagram
Physics-based modeling provides predictive understanding of selectively promiscuous substrate binding by Hsp70 chaperones | PLOS Computational Biology

Molecules | Free Full-Text | Substrate Binding Switches the Conformation at the Lynchpin Site in the Substrate-Binding Domain of Human Hsp70 to Enable Allosteric Interdomain Communication
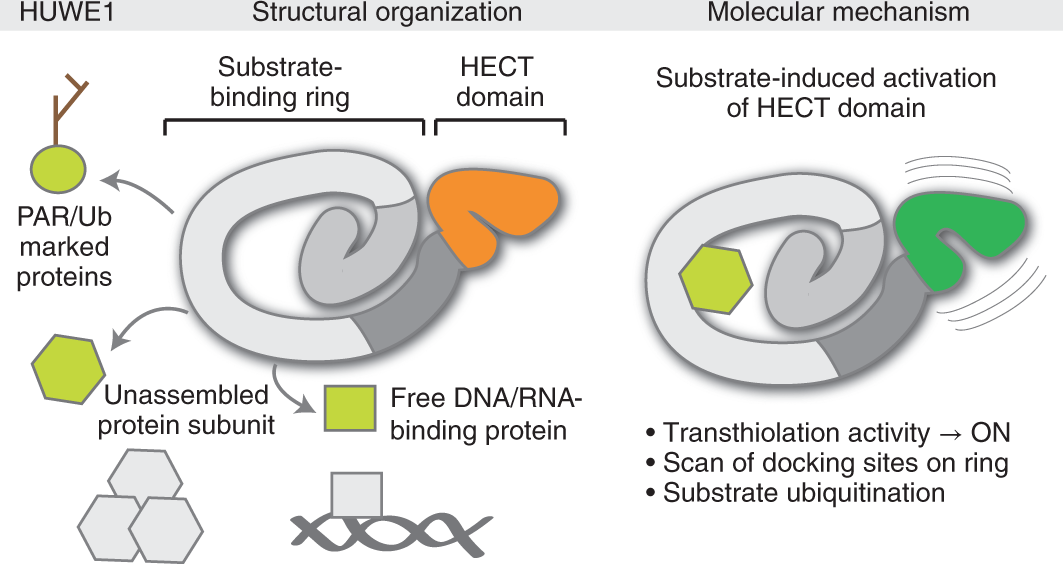
HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain | Nature Chemical Biology
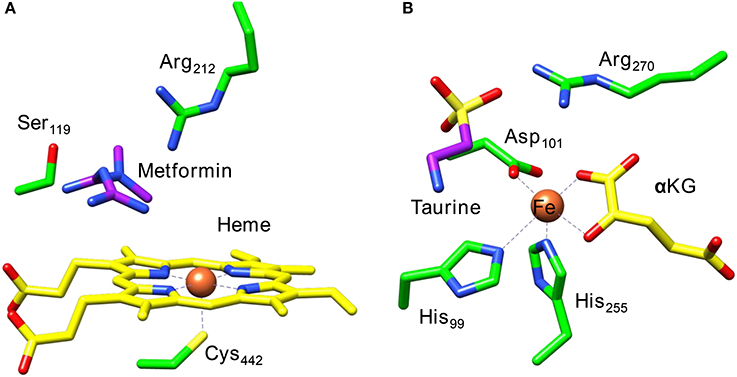
Frontiers | How Are Substrate Binding and Catalysis Affected by Mutating Glu127 and Arg161 in Prolyl-4-hydroxylase? A QM/MM and MD Study
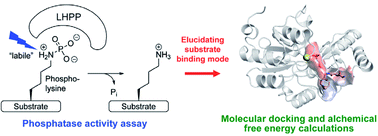
Combining free energy calculations with tailored enzyme activity assays to elucidate substrate binding of a phospho-lysine phosphatase - Chemical Science (RSC Publishing)

Application of cation-π interactions in enzyme-substrate binding: Design, synthesis, biological evaluation, and molecular dynamics insights of novel hydrophilic substrates for NQO1 - ScienceDirect

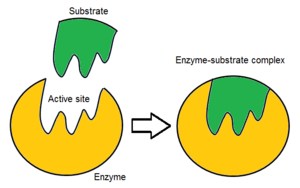

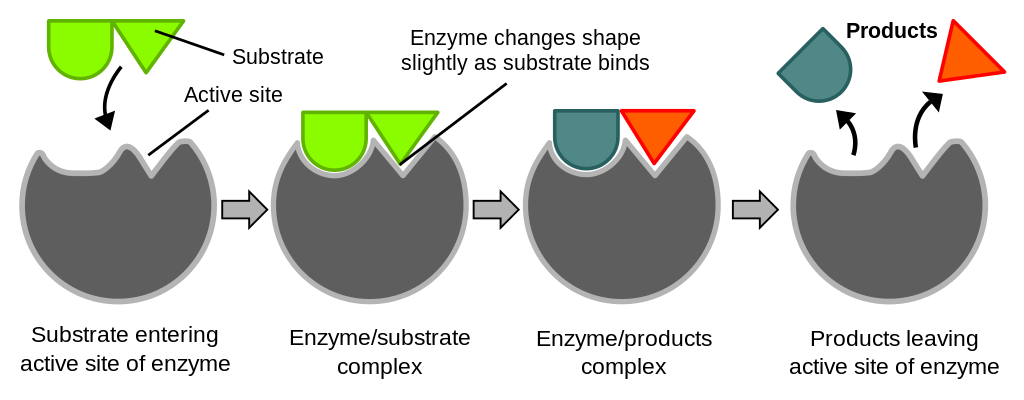
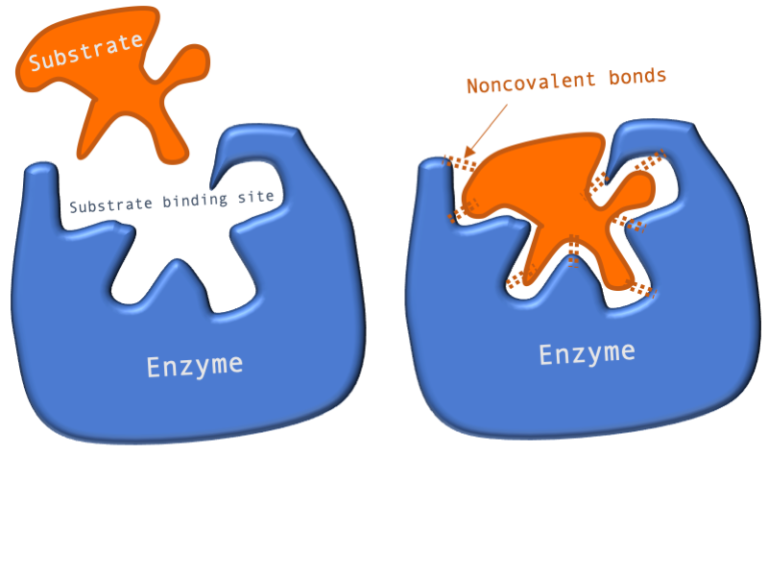


![2: Enzyme-substrate binding [17] | Download Scientific Diagram 2: Enzyme-substrate binding [17] | Download Scientific Diagram](https://www.researchgate.net/publication/307965030/figure/fig1/AS:643934288089090@1530537306480/Enzyme-substrate-binding-17.png)